Poisson 2D Example on Circular Domain
This example demonstrates how to solve a Poisson equation in 2D on a
circular domain using the fastvpinns package.
All the necessary files can be found in the examples folder of the fastvpinns GitHub repository
The Poisson equation is given by
where (\(\Omega\)) is the circular domain and (f) is the source term. The boundary conditions are given by
For this problem, the parameters are
The exact solution is given by
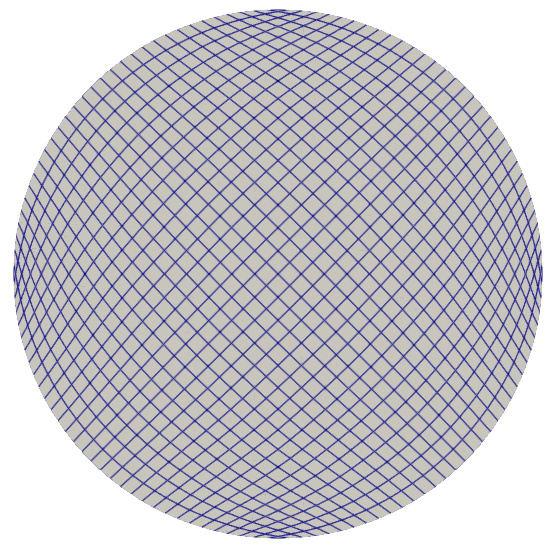
Computational Domain
The computational domain is a circular domain with radius 1 centered at (0, 0).
Contents
Steps to run the code
To run the code, execute the following command:
python3 main_poisson2d.py input.yaml
Example File
The example file, sin_cos.py, hosts all the details about the bilinear parameters for the
PDE, boundary conditions, source term, and the exact solution.
Defining the boundary conditions
The function circle_boundary returns the boundary value for a given
component of the boundary. The function get_boundary_function_dict
returns a dictionary of boundary functions. The key of the dictionary is
the boundary id and the value is the boundary function. The function
get_bound_cond_dict returns a dictionary of boundary conditions.
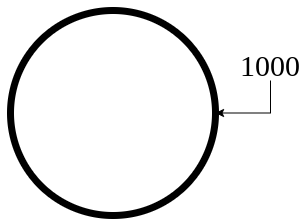
For externally created geometries from gmsh, the user needs to provide the physical tag for the boundaries present in the geometry. In our case, we have used 1000 to define the circular boundary in mesh file.
Note : As of now, only Dirichlet boundary conditions are supported.
def circle_boundary(x, y):
"""
This function will return the boundary value for given component of a boundary
"""
omegaX = 2.0 * np.pi
omegaY = 2.0 * np.pi
return -1.0 * np.sin(omegaX * x) * np.sin(omegaY * y)
def get_boundary_function_dict():
"""
This function will return a dictionary of boundary functions
"""
return {1000: circle_boundary}
def get_bound_cond_dict():
"""
This function will return a dictionary of boundary conditions
"""
return {1000: "dirichlet"}
Defining the source term
The function rhs returns the value of the source term at a given
point.
def rhs(x, y):
"""
This function will return the value of the rhs at a given point
"""
omegaX = 2.0 * np.pi
omegaY = 2.0 * np.pi
f_temp = -2.0 * (omegaX**2) * (np.sin(omegaX * x) * np.sin(omegaY * y))
return f_temp
Defining the exact solution
The function exact_solution returns the value of the exact solution
at a given point.
def exact_solution(x, y):
"""
This function will return the exact solution at a given point
"""
omegaX = 2.0 * np.pi
omegaY = 2.0 * np.pi
val = -1.0 * np.sin(omegaX * x) * np.sin(omegaY * y)
return val
Defining the bilinear form
The function get_bilinear_params_dict returns a dictionary of
bilinear parameters. The dictionary contains the values of the
parameter \(\varepsilon\) (diffusion coefficient).
Note : If any of the bilinear parameters are not present in the dictionary (for the cd2d model), then the code will throw an error.
def get_bilinear_params_dict():
"""
This function will return a dictionary of bilinear parameters
"""
eps = 1.0
return {"eps": eps}
Input File
This is the file that contains all the details about the problem. The input file is in the YAML format. The input file for this example is given below. The contents of the yaml files are as follows
Experimentation parameters
Defines the output path where the results will be saved.
experimentation:
output_path: "output/helmholtz/1"
Geometry parameters
It contains the details about the geometry of the domain. The mesh
generation method can be either “internal” or “external”. If the mesh
generation method is “internal”, then the internal_mesh_params are
used to generate the mesh. If the mesh generation method is “external”,
then the mesh is read from the file specified in the mesh_file
parameter.
In this case, we will use an external mesh. The mesh
../meshes/circle_quad.meshis generated using the Gmsh software. The mesh needs to have physical elements defined for the boundary. In this case, the physical element is defined as 1000 (which is defined in thecircle_boundaryfunction in thesin_cos.pyfile).exact_solution_generationis set to “internal” which means that the exact solution is generated using theexact_solutionfunction in thesin_cos.pyfile.mesh_typeis set to “quadrilateral” which means that the mesh is a quadrilateral mesh. (Note: As of now, only quadrilateral meshes are supported.)boundary_refinement_levelis set to 4 which means that the boundary is refined 4 times. (i.e), when the mesh is read, only the boundary points of an edge in quadrilateral mesh are read. This refinement will refine the boundary points to get more boundary points within the edge.boundary_sampling_methodis set to “uniform” which means that the boundary points are sampled using the “uniform” method. (Note: As of now, use only uniform sampling.)generate_mesh_plotis set to True which means that the mesh plot is generated and saved in the output directory.
geometry:
mesh_generation_method: "external" # Method for generating the mesh. Can be "internal" or "external".
generate_mesh_plot: True # Flag indicating whether to generate a plot of the mesh.
# internal mesh generated quadrilateral mesh, depending on the parameters specified below.
internal_mesh_params: # Parameters for internal mesh generation method.
x_min: 0 # Minimum x-coordinate of the domain.
x_max: 1 # Maximum x-coordinate of the domain.
y_min: 0 # Minimum y-coordinate of the domain.
y_max: 1 # Maximum y-coordinate of the domain.
n_cells_x: 4 # Number of cells in the x-direction.
n_cells_y: 4 # Number of cells in the y-direction.
n_boundary_points: 400 # Number of boundary points.
n_test_points_x: 100 # Number of test points in the x-direction.
n_test_points_y: 100 # Number of test points in the y-direction.
exact_solution:
exact_solution_generation: "internal" # whether the exact solution needs to be read from external file.
exact_solution_file_name: "" # External solution file name.
mesh_type: "quadrilateral" # Type of mesh. Can be "quadrilateral" or other supported types.
external_mesh_params: # Parameters for external mesh generation method.
mesh_file_name: "../meshes/circle_quad.mesh" # Path to the external mesh file (should be a .mesh file).
boundary_refinement_level: 4 # Level of refinement for the boundary.
boundary_sampling_method: "lhs" # Method for sampling the boundary. Can be "uniform" or "lhs".
Finite Element Space parameters
This section contains the details about the finite element spaces.
fe:
fe_order: 2 # Order of the finite element basis functions.
fe_type: "legendre" # Type of finite element basis functions. Can be "jacobi" or other supported types.
quad_order: 3 # Order of the quadrature rule.
quad_type: "gauss-jacobi" # Type of quadrature rule. Can be "gauss-jacobi" or other supported types.
Here the fe_order is set to 2 which means it has 2 basis functions
in each direction. The quad_order is set to 3 which means it uses a
3-points in each direction for the quadrature rule. The supported
quadrature rules are “gauss-jacobi” and “gauss-legendre”. In this
version of code, both “jacobi” and “legendre” refer to the same basis
functions (to maintain backward compatibility). The basis functions are
special type of Jacobi polynomials defined by
, where J n is the nth Jacobi polynomial.
PDE beta parameters
This value provides the beta values for the Dirichlet boundary conditions. The beta values are the multipliers that are used to multiply the boundary losses. The total loss is calculated as the sum of the PDE loss and the Dirichlet boundary loss, weighted by the beta values:
pde:
beta: 10 # Parameter for the PDE.
Model parameters
The model section contains the details about the dense model to be used.
The model architecture is given by the
model_architectureparameter.The activation function used in the model is given by the
activationparameter.The
epochsparameter is the number of training epochs.The
dtypeparameter is the data type used for computations.The
learning_ratesection contains the parameters for learning rate scheduling.The
initial_learning_rateparameter is the initial learning rate.The
use_lr_schedulerparameter is a flag indicating whether to use the learning rate scheduler.The
decay_stepsparameter is the number of steps between each learning rate decay.The
decay_rateparameter is the decay rate for the learning rate.The
staircaseparameter is a flag indicating whether to use the staircase decay.
Any parameter which are not mentioned above are archived parameters,
which are not used in the current version of the code (like
use_attention, set_memory_growth).
model:
model_architecture: [2, 30, 30, 30, 1] # Architecture of the neural network model.
activation: "tanh" # Activation function used in the neural network.
use_attention: False # Flag indicating whether to use attention mechanism in the model.
epochs: 5000 # Number of training epochs.
dtype: "float32" # Data type used for computations.
set_memory_growth: False # Flag indicating whether to set memory growth for GPU.
learning_rate: # Parameters for learning rate scheduling.
initial_learning_rate: 0.001 # Initial learning rate.
use_lr_scheduler: False # Flag indicating whether to use learning rate scheduler.
decay_steps: 1000 # Number of steps between each learning rate decay.
decay_rate: 0.99 # Decay rate for the learning rate.
staircase: False # Flag indicating whether to use staircase decay.
Logging parameters
update_console_output defines the epochs at which you need to log
parameters like loss, time taken, etc.
logging:
update_console_output: 2000
Main File
This file contains the main code to solve the Poisson equation in 2D on
a circular domain. The code reads the input file, sets up the problem,
and solves the Poisson equation using the fastvpinns package.
Importing the required libraries
The following libraries are imported in the main file.
import numpy as np
import pandas as pd
import pytest
import tensorflow as tf
from pathlib import Path
from tqdm import tqdm
import yaml
import sys
import copy
from tensorflow.keras import layers
from tensorflow.keras import initializers
from rich.console import Console
import copy
import time
Imports from fastvpinns
The following imports are used from the fastvpinns package.
Imports the geometry module from the
fastvpinnspackage, which contains theGeometry_2Dclass responsible for setting up the geometry of the domain.
from fastvpinns.Geometry.geometry_2d import Geometry_2D
Imports the fespace module from the
fastvpinnspackage, which contains theFEclass responsible for setting up the finite element spaces.
from fastvpinns.FE.fespace2d import Fespace2D
Imports the datahandler module from the
fastvpinnspackage, which contains theDataHandlerclass responsible for handling and converting the data to necessary shape for training purposes
from fastvpinns.DataHandler.datahandler import DataHandler
Imports the model module from the
fastvpinnspackage, which contains theModelclass responsible for training the neural network model.
from fastvpinns.Model.model import DenseModel
Import the Loss module from the
fastvpinnspackage, which contains the loss function of the PDE to be solved in tensor form.
from fastvpinns.physics.helmholtz2d import pde_loss_helmholtz
Import additional functionalities from the
fastvpinnspackage.
from fastvpinns.utils.plot_utils import plot_contour, plot_loss_function, plot_test_loss_function
from fastvpinns.utils.compute_utils import compute_errors_combined
from fastvpinns.utils.print_utils import print_table
Reading the input file
The input file is read using the yaml library.
if len(sys.argv) != 2:
print("Usage: python main.py <input file>")
sys.exit(1)
# Read the YAML file
with open(sys.argv[1], 'r') as f:
config = yaml.safe_load(f)
Reading all input parameters
# Extract the values from the YAML file
i_output_path = config['experimentation']['output_path']
i_mesh_generation_method = config['geometry']['mesh_generation_method']
i_generate_mesh_plot = config['geometry']['generate_mesh_plot']
i_mesh_type = config['geometry']['mesh_type']
i_x_min = config['geometry']['internal_mesh_params']['x_min']
i_x_max = config['geometry']['internal_mesh_params']['x_max']
i_y_min = config['geometry']['internal_mesh_params']['y_min']
i_y_max = config['geometry']['internal_mesh_params']['y_max']
i_n_cells_x = config['geometry']['internal_mesh_params']['n_cells_x']
i_n_cells_y = config['geometry']['internal_mesh_params']['n_cells_y']
i_n_boundary_points = config['geometry']['internal_mesh_params']['n_boundary_points']
i_n_test_points_x = config['geometry']['internal_mesh_params']['n_test_points_x']
i_n_test_points_y = config['geometry']['internal_mesh_params']['n_test_points_y']
i_exact_solution_generation = config['geometry']['exact_solution']['exact_solution_generation']
i_exact_solution_file_name = config['geometry']['exact_solution']['exact_solution_file_name']
i_mesh_file_name = config['geometry']['external_mesh_params']['mesh_file_name']
i_boundary_refinement_level = config['geometry']['external_mesh_params'][
'boundary_refinement_level'
]
i_boundary_sampling_method = config['geometry']['external_mesh_params'][
'boundary_sampling_method'
]
i_fe_order = config['fe']['fe_order']
i_fe_type = config['fe']['fe_type']
i_quad_order = config['fe']['quad_order']
i_quad_type = config['fe']['quad_type']
i_model_architecture = config['model']['model_architecture']
i_activation = config['model']['activation']
i_use_attention = config['model']['use_attention']
i_epochs = config['model']['epochs']
i_dtype = config['model']['dtype']
if i_dtype == "float64":
i_dtype = tf.float64
elif i_dtype == "float32":
i_dtype = tf.float32
else:
print("[ERROR] The given dtype is not a valid tensorflow dtype")
raise ValueError("The given dtype is not a valid tensorflow dtype")
i_set_memory_growth = config['model']['set_memory_growth']
i_learning_rate_dict = config['model']['learning_rate']
i_beta = config['pde']['beta']
i_update_console_output = config['logging']['update_console_output']
All the variables which are named with the prefix i_ are input
parameters which are read from the input file.
Setup the geometry
Obtain the boundary condition and boundary values from the
sin_cos.py file and initialise the Geometry_2D class. After that
use the domain.read_mesh functionality to read the external mesh
file.
cells, boundary_points = domain.read_mesh(
i_mesh_file_name,
i_boundary_refinement_level,
i_boundary_sampling_method,
refinement_level=1,
)
Setup fespace
Initialise the Fespace2D class with the required parameters.
fespace = Fespace2D(
mesh=domain.mesh,
cells=cells,
boundary_points=boundary_points,
cell_type=domain.mesh_type,
fe_order=i_fe_order,
fe_type=i_fe_type,
quad_order=i_quad_order,
quad_type=i_quad_type,
fe_transformation_type="bilinear",
bound_function_dict=bound_function_dict,
bound_condition_dict=bound_condition_dict,
forcing_function=rhs,
output_path=i_output_path,
generate_mesh_plot=i_generate_mesh_plot,
)
Setup datahandler
Initialise the DataHandler class with the required parameters.
datahandler = DataHandler2D(fespace, domain, dtype=i_dtype)
Setup model
Setup the necessary parameters for the model and initialise the Model
class. Before that fill the params dictionary with the required
parameters.
model = DenseModel(
layer_dims=i_model_architecture,
learning_rate_dict=i_learning_rate_dict,
params_dict=params_dict,
loss_function=pde_loss_cd2d,
input_tensors_list=[datahandler.x_pde_list, train_dirichlet_input, train_dirichlet_output],
orig_factor_matrices=[
datahandler.shape_val_mat_list,
datahandler.grad_x_mat_list,
datahandler.grad_y_mat_list,
],
force_function_list=datahandler.forcing_function_list,
tensor_dtype=i_dtype,
use_attention=i_use_attention,
activation=i_activation,
hessian=False,
)
Pre-train setup
test_points = domain.get_test_points()
print(f"[bold]Number of Test Points = [/bold] {test_points.shape[0]}")
y_exact = exact_solution(test_points[:, 0], test_points[:, 1])
# plot the exact solution
num_epochs = i_epochs # num_epochs
progress_bar = tqdm(
total=num_epochs,
desc='Training',
unit='epoch',
bar_format="{l_bar}{bar:40}{r_bar}{bar:-10b}",
colour="green",
ncols=100,
)
loss_array = [] # total loss
test_loss_array = [] # test loss
time_array = [] # time per epoc
# beta - boundary loss parameters
beta = tf.constant(i_beta, dtype=i_dtype)
Here the exact solution is being read from the external file. The external solution at the test points is computed by FEM and stored in a csv file. This sets up the test points and the exact solution. The progress bar is initialised and the loss arrays are set up. The beta value is set up as a constant tensor.
Training
for epoch in range(num_epochs):
# Train the model
batch_start_time = time.time()
loss = model.train_step(beta=beta, bilinear_params_dict=bilinear_params_dict)
elapsed = time.time() - batch_start_time
# print(elapsed)
time_array.append(elapsed)
loss_array.append(loss['loss'])
This train_step function trains the model for one epoch and returns
the loss. The loss is appended to the loss array. Then for every epoch
where
(epoch + 1) % i_update_console_output == 0 or epoch == num_epochs - 1:
y_pred = model(test_points).numpy()
y_pred = y_pred.reshape(-1)
error = np.abs(y_exact - y_pred)
# get errors
(
l2_error,
linf_error,
l2_error_relative,
linf_error_relative,
l1_error,
l1_error_relative,
) = compute_errors_combined(y_exact, y_pred)
loss_pde = float(loss['loss_pde'].numpy())
loss_dirichlet = float(loss['loss_dirichlet'].numpy())
total_loss = float(loss['loss'].numpy())
# Append test loss
test_loss_array.append(l1_error)
solution_array = np.c_[y_pred, y_exact, np.abs(y_exact - y_pred)]
domain.write_vtk(
solution_array,
output_path=i_output_path,
filename=f"prediction_{epoch+1}.vtk",
data_names=["Sol", "Exact", "Error"],
)
console.print(f"\nEpoch [bold]{epoch+1}/{num_epochs}[/bold]")
console.print("[bold]--------------------[/bold]")
console.print("[bold]Beta : [/bold]", beta.numpy(), end=" ")
console.print(
f"Variational Losses || Pde Loss : [red]{loss_pde:.3e}[/red] Dirichlet Loss : [red]{loss_dirichlet:.3e}[/red] Total Loss : [red]{total_loss:.3e}[/red]"
)
console.print(
f"Test Losses || L1 Error : {l1_error:.3e} L2 Error : {l2_error:.3e} Linf Error : {linf_error:.3e}"
)
We will compute all the test errors and write the solution to a vtk file for a complex mesh. Further, the console output will be printed with the loss values and the test errors.
Post Training
# Save the model
model.save_weights(str(Path(i_output_path) / "model_weights"))
solution_array = np.c_[y_pred, y_exact, np.abs(y_exact - y_pred)]
domain.write_vtk(
solution_array,
output_path=i_output_path,
filename=f"prediction_{epoch+1}.vtk",
data_names=["Sol", "Exact", "Error"],
)
# print the Error values in table
print_table(
"Error Values",
["Error Type", "Value"],
[
"L2 Error",
"Linf Error",
"Relative L2 Error",
"Relative Linf Error",
"L1 Error",
"Relative L1 Error",
],
[l2_error, linf_error, l2_error_relative, linf_error_relative, l1_error, l1_error_relative],
)
# print the time values in table
print_table(
"Time Values",
["Time Type", "Value"],
[
"Time per Epoch(s) - Median",
"Time per Epoch(s) IQR-25% ",
"Time per Epoch(s) IQR-75% ",
"Mean (s)",
"Epochs per second",
"Total Train Time",
],
[
np.median(time_array),
np.percentile(time_array, 25),
np.percentile(time_array, 75),
np.mean(time_array),
int(i_epochs / np.sum(time_array)),
np.sum(time_array),
],
)
# save all the arrays as numpy arrays
np.savetxt(str(Path(i_output_path) / "loss_function.txt"), np.array(loss_array))
np.savetxt(str(Path(i_output_path) / "prediction.txt"), y_pred)
np.savetxt(str(Path(i_output_path) / "exact.txt"), y_exact)
np.savetxt(str(Path(i_output_path) / "error.txt"), error)
np.savetxt(str(Path(i_output_path) / "time_per_epoch.txt"), np.array(time_array))
This part of the code saves the model weights, writes the solution to a vtk file, prints the error values in a table, prints the time values in a table, and saves all the arrays as numpy arrays.
Save the outputs
All the outputs will be saved in the output directory specified in the input file. The output directory will contain the following files:
prediction_{epoch}.vtk : The solution file for each epoch.
loss_function.txt : The loss function values for each epoch.
prediction.txt : The predicted values at last epoch at the test points.
exact.txt : The exact values at last epoch at the test points.
error.txt : The error values at last epoch at the test points.
time_per_epoch.txt : The time taken for each epoch.
Solution Plots
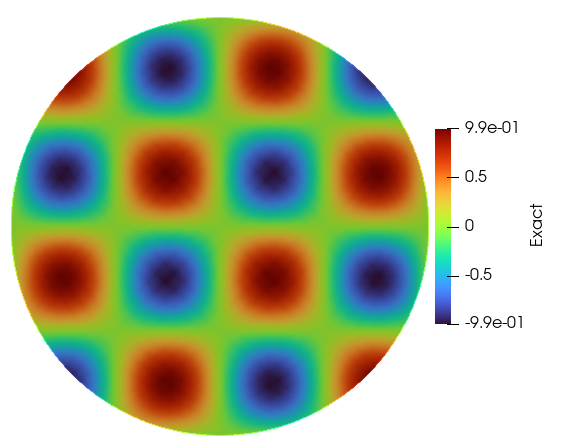
Exact Solution
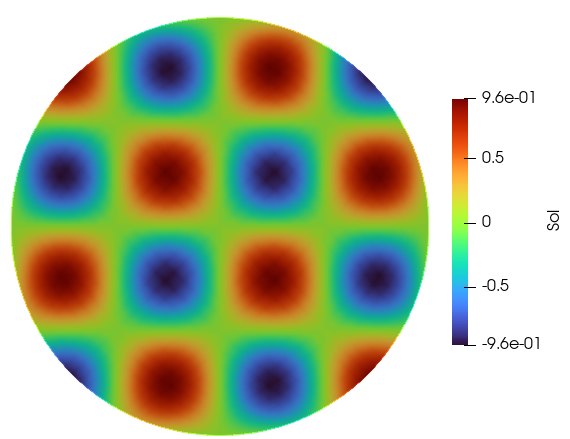
Predicted Solution
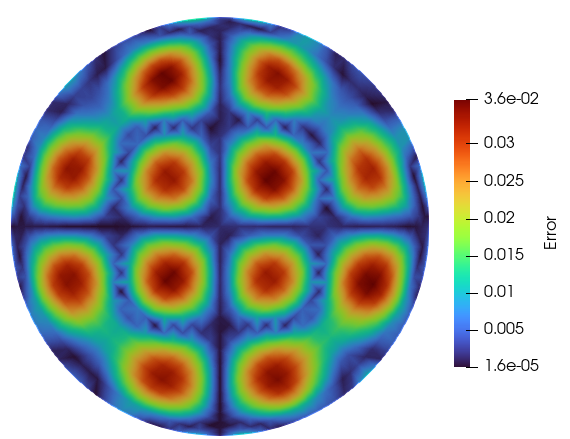
Error